Peptide Hits Result Processor
Converts a MSGF+ TSV file, X!Tandem results file (XML format), or a SEQUEST Synopsis/First Hits file to a series of tab-delimited text files summarizing the results.
Description
Parallel files are created containing sequence information, modification details, and protein information. The user can optionally provide a modification definition file that specifies the symbol to use for each modification mass.
When parsing X!Tandem data, the program will insert modification symbols into the peptide sequences for modified peptides.
The “Data Extraction and Analysis for LC-MS Based Proteomics” sessions at the 2007 and 2008 US HUPO conferences discussed the use of the Peptide Hit Results Processor for processing X!Tandem results. PDF files of the slides presented are available for download as a 5 MB PDF file (2007) and a 6.5 MB PDF file (2008).
Note that you can generate SEQUEST Synopsis/First Hits files from SEQUEST .Out files using the Peptide File Extractor.
Downloads
Software Instructions
Run PeptideHitResultsProcessor_Installer.msi to install the application. This is a command-line (console) application and thus does not have a GUI. For more information on using command line applications, see the Command Line Application help page.
Compressed file MSGFPlus_Example_Data.zip contains example data files and additional information on running MSGF+. You can find this file in the application folder (the folder with PeptideHitResultsProcRunner.exe). Simply unzip this file to a local directory on your computer to see the example data files. This file is also available for download from this web page.
-
X!Tandem Synopsis File Columns Example X!Tandem XML Results File

Example Tab-Delimited Text Output File (X!Tandem Results)
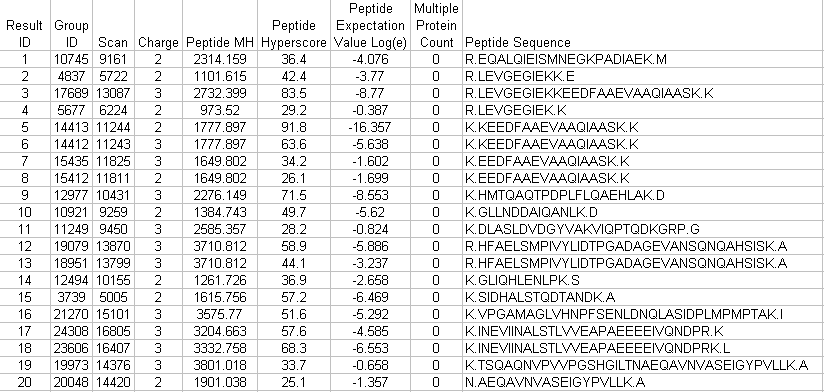
-
MSGF+ Synopsis File Columns Example Tab-Delimited Text Output File (MSGF+)
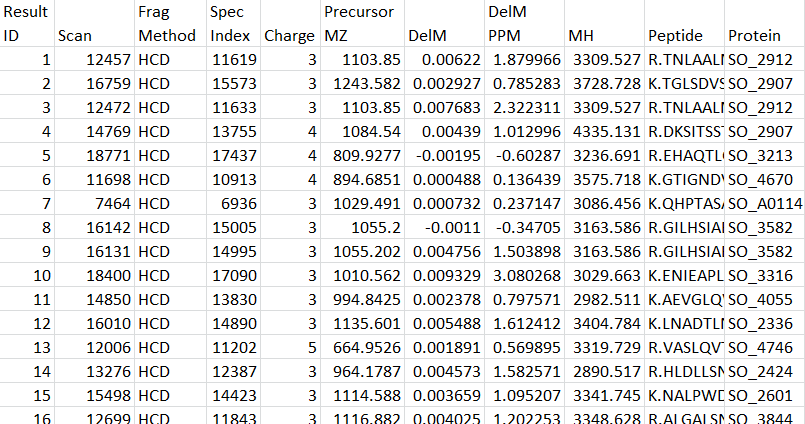
Acknowledgment
All publications that utilize this software should provide appropriate acknowledgement to PNNL and the PHRP GitHub repository. However, if the software is extended or modified, then any subsequent publications should include a more extensive statement, as shown in the Readme file for the given application or on the website that more fully describes the application.
Disclaimer
These programs are primarily designed to run on Windows machines. Please use them at your own risk. This material was prepared as an account of work sponsored by an agency of the United States Government. Neither the United States Government nor the United States Department of Energy, nor Battelle, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness or any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights.
Portions of this research were supported by the NIH National Center for Research Resources (Grant RR018522), the W.R. Wiley Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy’s Office of Biological and Environmental Research and located at PNNL), and the National Institute of Allergy and Infectious Diseases (NIH/DHHS through interagency agreement Y1-AI-4894-01). PNNL is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC05-76RL0 1830.
We would like your feedback about the usefulness of the tools and information provided by the Resource. Your suggestions on how to increase their value to you will be appreciated. Please e-mail any comments to proteomics@pnl.gov