PNNL PreProcessor
Ion mobility-mass spectrometry (IM-MS) provides an increasingly popular platform for analyzing complex samples due to its separation power and ability to differentiate structural isomers, which are difficult to resolve using conventional LC-MS systems. Here we provide a software tool with various algorithms and utilities to improve workflows using this technology: data compression and interpolation, ion mobility demultiplexing, multidimensional smoothing, noise filtering by low intensity threshold and spike removal, saturation repair and metadata export.
Description
In close collaboration between Pacific Northwest National Laboratory and Agilent Technologies, we have developed this user-friendly tool for Agilent MassHunter (.d) and UIMF mass spectrometry data files (MS-files) from drift tube (DT) and structure for lossless ion manipulations (SLIM) IM-MS platforms, to generate new MS-files in the same instrument data format with enhanced signal quality.
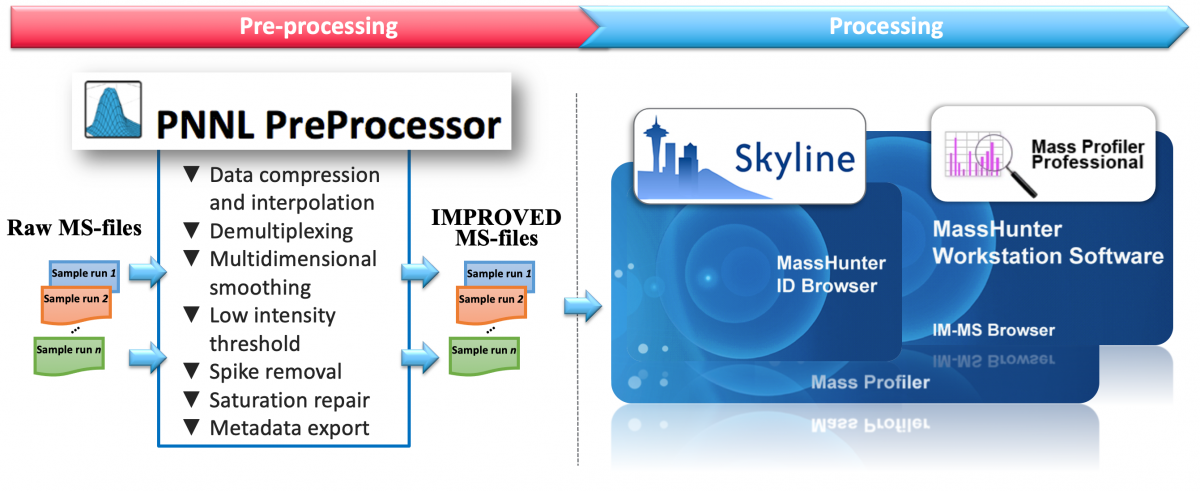
Software features:
- Command-line and graphical (workflow style) user interfaces.
- Single-click batch processing of multiple raw MS-files.
- Data compression (by frame and mobility) and filtering by retention time range.
- Data interpolation of the ion mobility dimension to improve the results of the HRdm demultiplexing and peak deconvolution strategy.
- Multidimensional smoothing of data and repair of saturated peaks.
- PNNL demultiplexing and artifact removal algorithm integrated. A new selectable pulse coverage percentage to increase sensitivity for low level signals.
- An algorithm to remove noise in form of ‘spikes’.
- Ion mobility MS with/without any separation: LC-IM-MS, solid phase extraction (e.g., RapidFire) IM-MS and direct infusion IM-MS.
- All Ions MS-files (data-independent acquisition) with alternating high/low collision energy fragmentation.
- Exporting metadata information of ion mobility frames (e.g., field, pressure, temperature) and MS actuals to text files.
- Multidimensional smoothing for non-ion mobility TOF-MS MS-files (e.g., produced by 6530, 6540, 6545 and 6550 Agilent instruments).
- Conversion of arrival time to CCS in the raw data for SLIM.
- Conversion of arrival time to retention time: single frame “IMMS” data to “LCMS” format.
New features:
- Polygon extraction is available for batch processing in Step 4a. (Requires a .m method file from IM-MS Browser)
- The following features require additional IM-MS Browser dlls and are not available if they are missing. Instructions to obtain these dlls are in the User Guide.
- IM to DDA conversion of All Ions IM/MS mobility aligned fragmentation data can now call IMFE from within the PNNL PreProcessor to generate a precursor feature list.
- Batch/bulk CCS Calibration has been added as a capability; this processing calls IMFE from within the PNNL PreProcessor for feature finding.
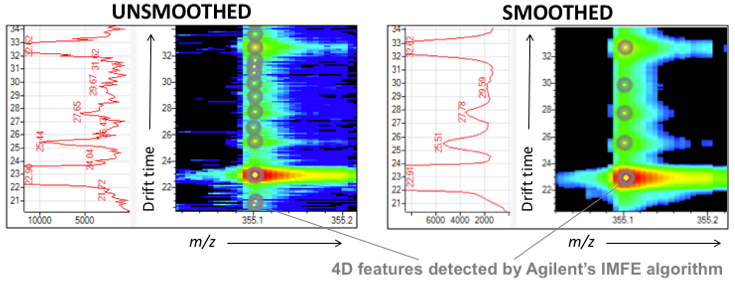
Smoothing removes artifacts in jagged peaks, which are common in low-abundance ions. Real signals are enhanced, and variations in abundance, elution and mobility/collisional-cross-section are reduced.
Disclaimer: the saturation repair software may produce incorrect results for ions with highly convoluted elution/mobility profiles caused by interferences. Please use the current version at your own risk and check the output log files in each data file to verify the repairs made.
How to Cite
If you use the PreProcessor, please cite: Bilbao et al. A Preprocessing Tool for Enhanced Ion Mobility-Mass Spectrometry-Based Omics Workflows. Journal of Proteome Research 2021 https://doi.org/10.1021/acs.jproteome.1c00425.
If you use the IM-to-LC, IM-to-CCS, or IM-to-DDA conversions, please cite: Stow et al. Exploring Ion Mobility Mass Spectrometry Data File Conversions to Leverage Existing Tools and Enable New Workflows. Journal of the American Society for Mass Spectrometry 2024. https://pubs.acs.org/doi/full/10.1021/jasms.4c00220.
Downloads
- All downloads
- Software (as .zip or installer)
- User Guide
Requirements
- Supports MS-files in UIMF or Agilent MassHunter (.d) format (note: Bruker .d format will not work)
Hardware Requirements
- Minimum requirements:
- 8 GB memory
- Sufficient free hard disk space
- Recommended requirements:
- Quad core, 3.0 GHz processor
- 32 GB memory
- SSD (solid state drive) or large (>2 TB) hard drive for reading/writing the data files (provides a minor improvement)
- Demultiplexing recommended requirements:
- Significant performance improvements have been noted on high-core-count processors
- Processor with 8 or more cores and 3.5 GHz or faster speed
- 32 GB minimum for 8 processor cores, more for higher core counts
The hardware configuration used for development and testing, which provides reasonable performance:
- 8-core Intel Xeon W, at 3.7 or 3.9 GHz
- 32 GB memory
- SSD for the operating system, 4 TB hard drive for data
An example hardware configuration that has significant processing time improvements for demultiplexing:
- 24- or 32-core AMD Ryzen Threadripper processor, at 3.5 GHz or higher speeds
- 128 GB memory
- SSD for the operating system, 4 or 8 TB hard drive for data
Software Requirements
- Windows 7 (64-bit) or later Windows 64-bit operating system
- .NET Framework 4.7.2 or later (included with Windows 10 update 1803 and later releases, for earlier Windows you can download from Microsoft)
- Microsoft Visual C++ Runtime x64 (may already be installed, if the program doesn’t work then you can download vcredist_x64.exe from Microsoft - the version ‘Visual Studio 2013 (VC++ 12.0)’ is needed)
Acknowledgment
All publications that utilize this software should provide appropriate acknowledgement to PNNL and the PNNL-Preprocessor publication.
For questions about the possibility of extending or modifying the software please contact aivett.bilbao@pnnl.gov.
License / Disclaimer
Copyright 2024, Battelle Memorial Institute
Notice: These data were produced by Battelle Memorial Institute under Contract No. DE-AC05-76RL01830 with the Department of Energy. During the period of commercialization or such other time period specified by DOE, the Government is granted for itself and others acting on its behalf a nonexclusive, paid-up, irrevocable worldwide license in this data to reproduce, prepare derivative works, and perform publicly and display publicly, by or on behalf of the Government. Subsequent to that period, the Government is granted for itself and others acting on its behalf a nonexclusive, paid-up, irrevocable worldwide license in this data to reproduce, prepare derivative works, distribute copies to the public, perform publicly and display publicly, and to permit others to do so. The specific term of the license can be identified by inquiry made to Contractor or DOE. NEITHER THE UNITED STATES NOR THE UNITED STATES DEPARTMENT OF ENERGY, NOR ANY OF THEIR EMPLOYEES, MAKES ANY WARRANTY, EXPRESS OR IMPLIED, OR ASSUMES ANY LEGAL LIABILITY OR RESPONSIBILITY FOR THE ACCURACY, COMPLETENESS, OR USEFULNESS OF ANY DATA, APPARATUS, PRODUCT, OR PROCESS DISCLOSED, OR REPRESENTS THAT ITS USE WOULD NOT INFRINGE PRIVATELY OWNED RIGHTS.
License addendum
This software binary is provided free of charge (closed-source code due to restrictions from the instrument vendor on their proprietary formats).
However, the software binary depends on other software libraries which place further restrictions on its use and redistribution. By using PNNL-PreProcessor, you agree to comply with the restrictions imposed on you by the license agreements of the software libraries on which it depends: Agilent Technologies Mass Hunter Data Access Component Library (www.agilent.com).